Logistic Regression¶
Learning Objectives¶
- Logistic Regression
- Concepts of machine learning:
- Classification
- Linear separability, decision boundaries
- Cost function for classification
- Recap: Gradient descent
Problem Formulation¶
Classification¶
2-class problem:
$$ y \in \{ 0, 1\} $$- $y=1$: positive class, e.g. email: spam
- $y=0$: negative class, e.g. email: no spam
(regression: $y \in \mathbb R$)
Classification with linearer regression¶
Lineare Function $$ h_\theta(\vec{x}) = \vec \theta^T \cdot \vec{x} $$
Threshold for classification
- $h_{\Theta}(\vec{x}) \geq 0.5$ $\Rightarrow$ positive class
- $h_{\Theta}(\vec x) < 0.5 $ $\Rightarrow$ negative class
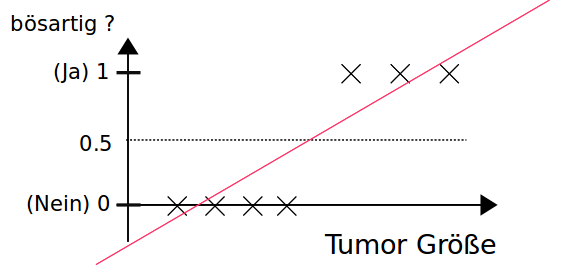
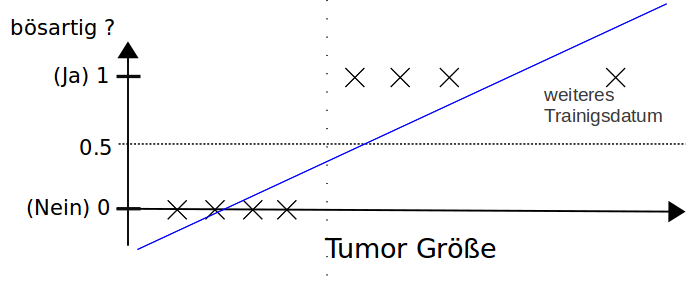
Using linear regression with a threshold for classification don't work properly.
Also note:
$h_{\Theta}(\vec x)$ can be greater than 1 or smaller than 0.
Logistic function (sigmoid)¶
We are looking for a function to squash the output:
$$ 0 \leq g(z) \leq 1 $$That holds for the logistic function (sigmoid):
$$ g(z) = \frac{1}{1 + \exp(-z)} $$Note:
$$ \frac{1}{1 + \exp(0)} = \frac{1}{2} $$import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
def sigmoid(z):
return 1/(1+np.exp(-z))
z=np.arange(-10.,10,0.1)
plt.plot(z, sigmoid(z), 'b-')
plt.ylim(-.1, 1.1)
plt.xlabel('z'); plt.ylabel('sigmoid(z)')
Logistic Regression Hypothesis¶
$$ h_\theta(\vec x) = g(\vec \theta^T \vec x) $$with the logistic Function $g(z)$:
$$ h_\theta(\vec x) = \frac{1}{1 + \exp(-\vec \theta^T \vec x)} $$Name 'logistic regression'¶
- Name part “logistic”: due to the use of the logistic function.
- Name part “regression” has historical reasons. But, it's not regression. It's classification!
Interpretation as probability¶
Discriminative model:
$$ h_\theta(\vec x) = p(y=1 \mid {\vec x}, \theta) $$Conditional probability of $y=1$ under the constraint ${\vec x}$ and parameters $\theta$.
e.g. $ h_\theta({\vec x}) = 0.8$: probability for a malign tumor is 80%.
Prediction according to the probabilities. Usually: \begin{align} h\theta({\vec x}) & \geq 0.5 \rightarrow y=1 \ h\theta({\vec x}) & < 0.5 \rightarrow y=0 \end{align}
Decision Boundary¶
Decision boundary by thesholding¶
with threshold value 0.5 we have:
\begin{align} \vec \theta^T \cdot \vec x & \geq 0 \Rightarrow h\theta({\bf x}) \geq 0.5 \Rightarrow y{predicted}=1 \ \vec \theta^T \cdot \vec x & < 0 \Rightarrow h\theta({\bf x}) < 0.5 \Rightarrow y{predicted}=0 \end{align}
Example 1: $$ \vec \theta^T \vec x = \theta_0 + \theta_1 x_1 = -6. + \frac{6}{5} x_1 $$
Decision Boundary: $\vec \theta^T \vec x = 0 $
$\Rightarrow x_1 = 5$
Prediction:
- $x_1 \geq 5 \Rightarrow y_{predict} = 1$
- $x_1 < 5 \Rightarrow y_{predict} = 0$
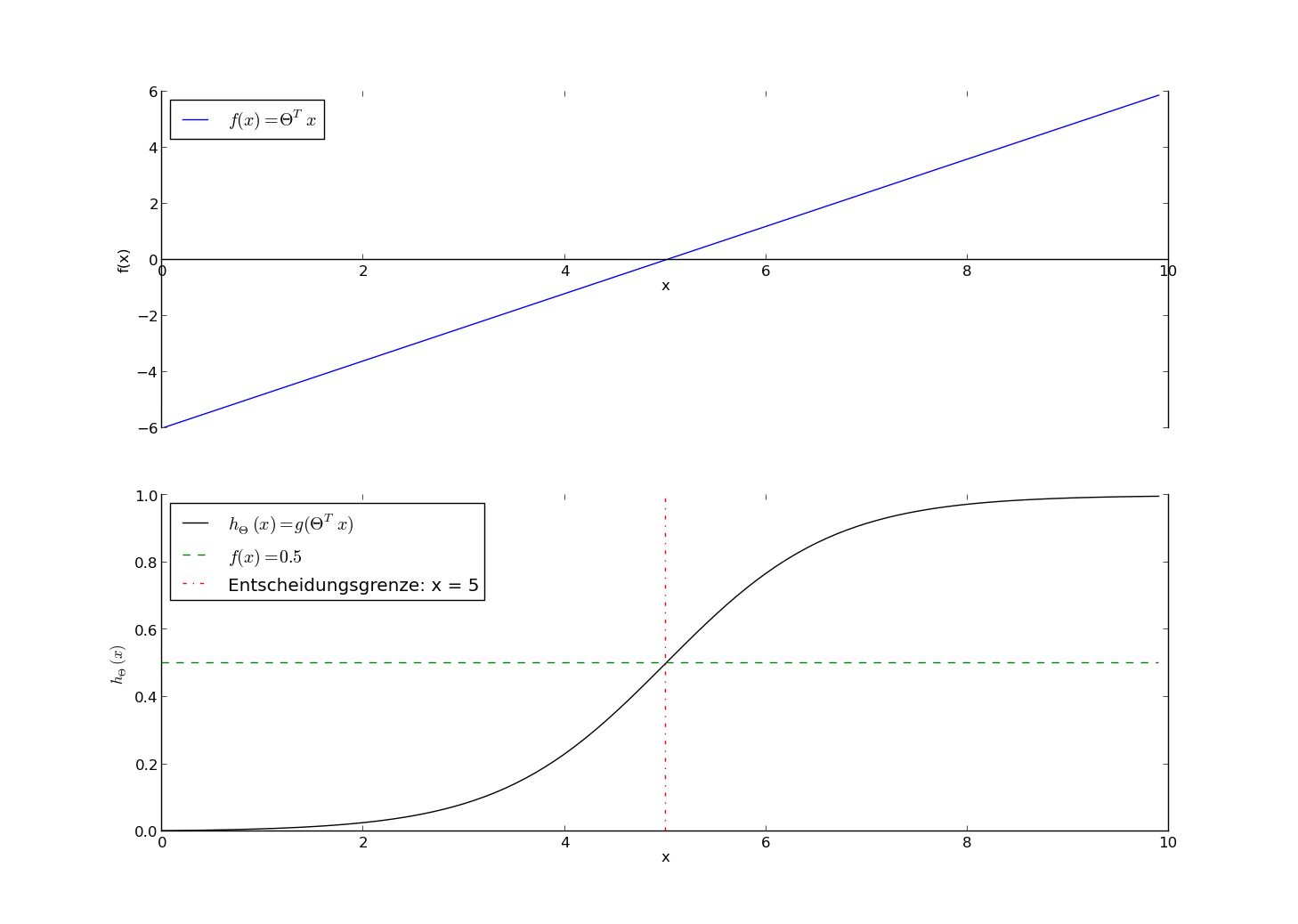
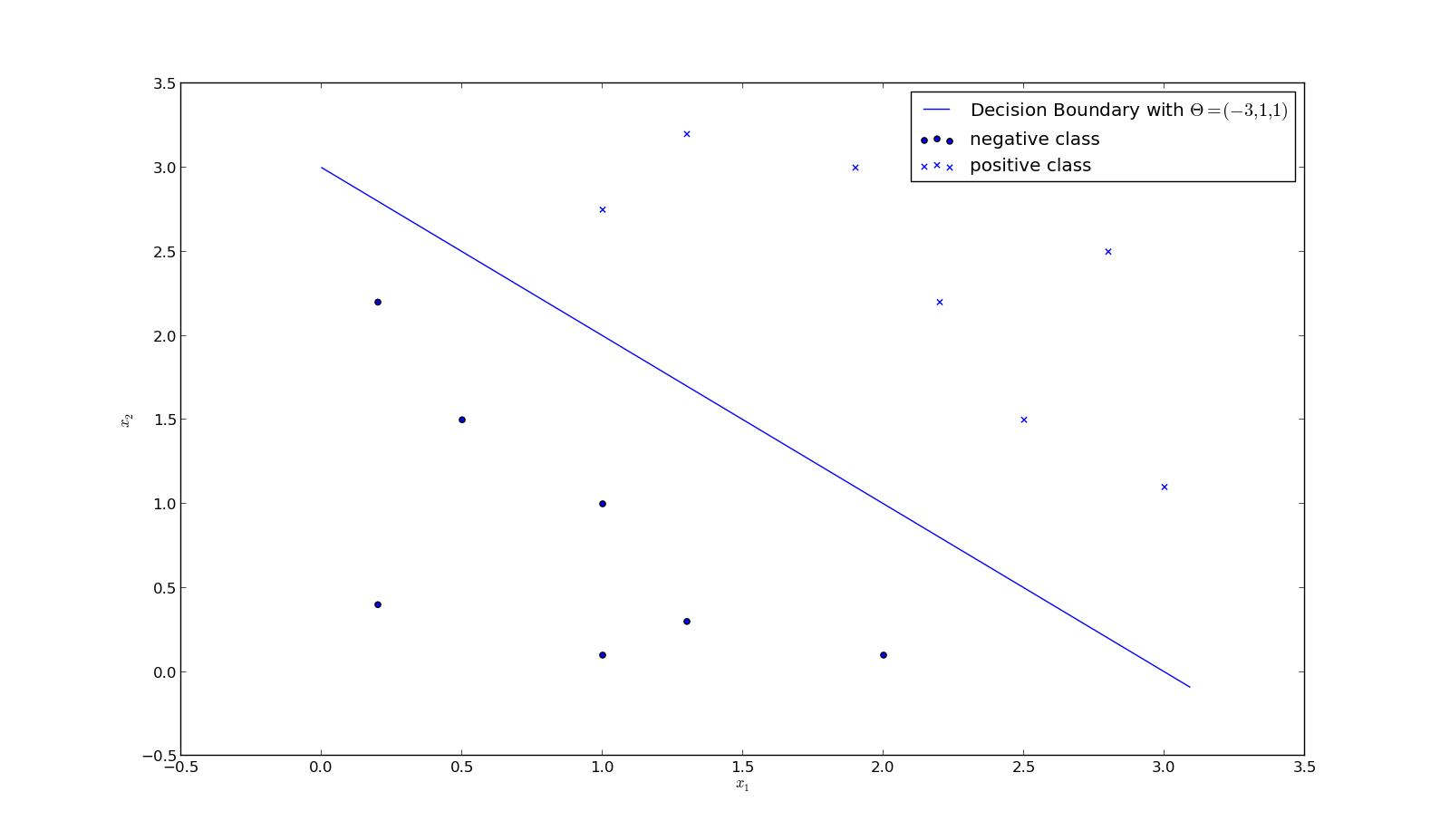
Example 2:
$$ \vec \theta^T \vec x = \theta_0 + \theta_1 x_1 + \theta_2 x_2 = 3 - x_1 - x_2 $$Decision boundary:
$\vec \theta^T \vec x = 0 $ $\Rightarrow x_2 = 3 - x_1$
Prediction:
- $x_1 + x_2 \geq 3 \Rightarrow y_{predict} = 1$
- $x_1 + x_2 < 3 \Rightarrow y_{predict} = 0$
Example: Iris data set (just 2 classed and two features)
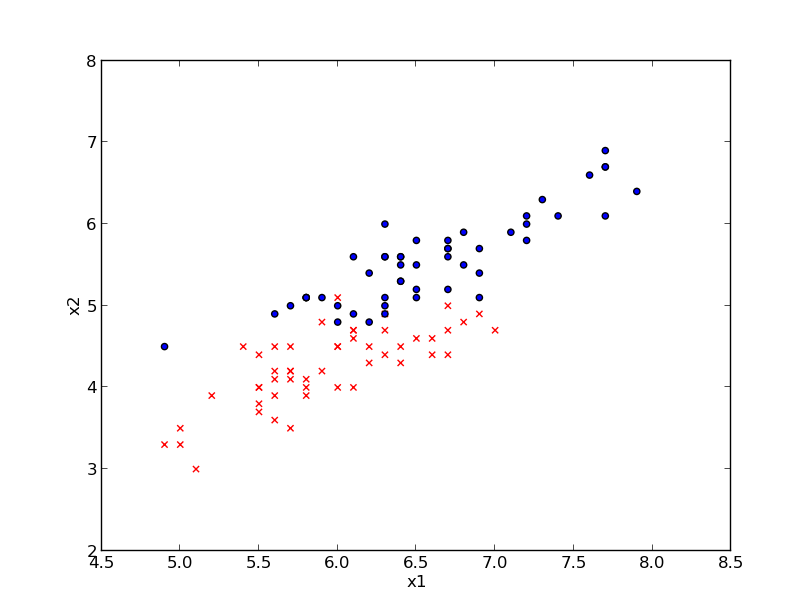
Classes: Iris-Versicolour (red), Iris-Virginica (blue)
Features: $x_1$: sepal length (cm) \hspace {1.5cm} $x_2$: petal length (cm)
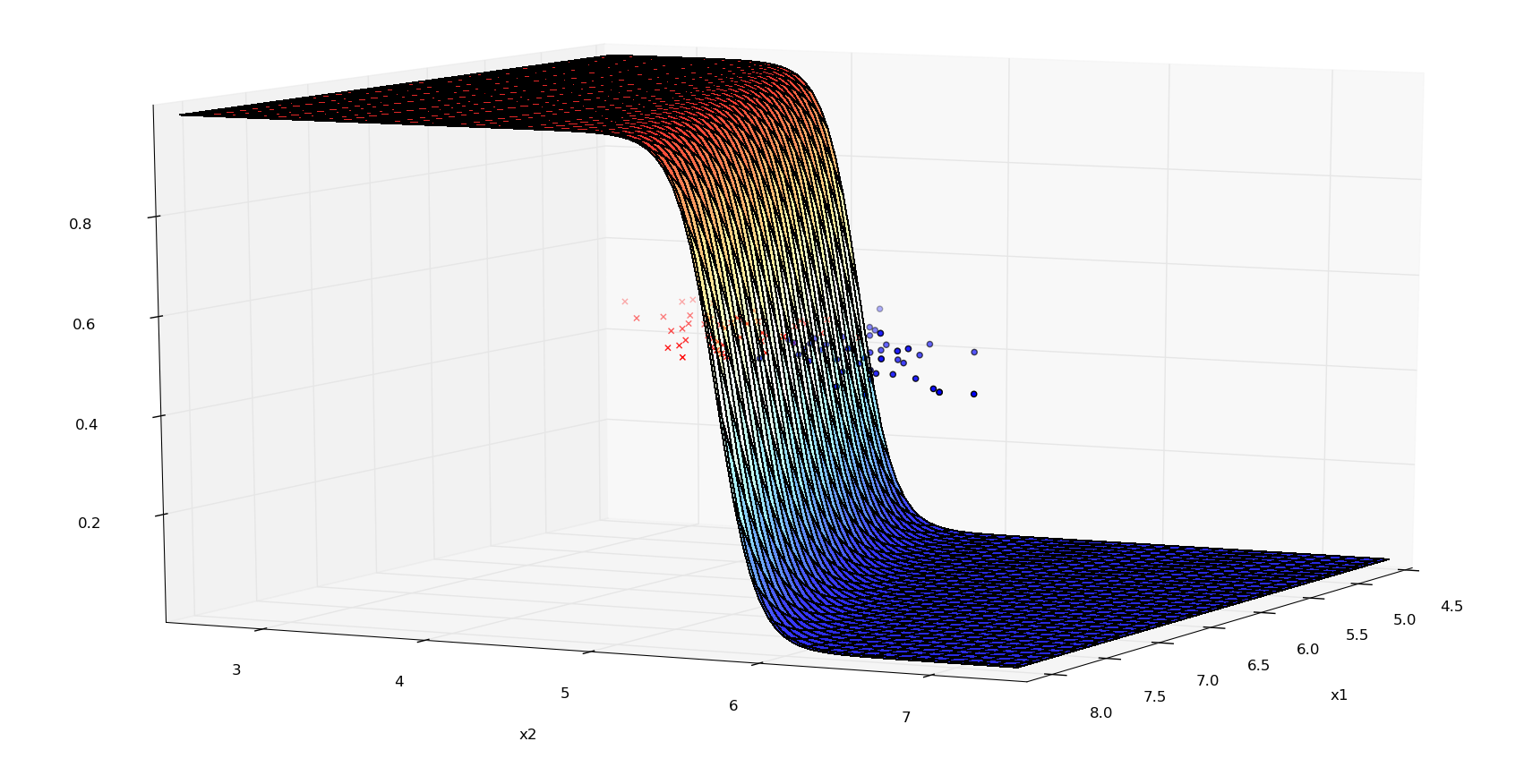
Probabilities (3d-plot) - Iris data:
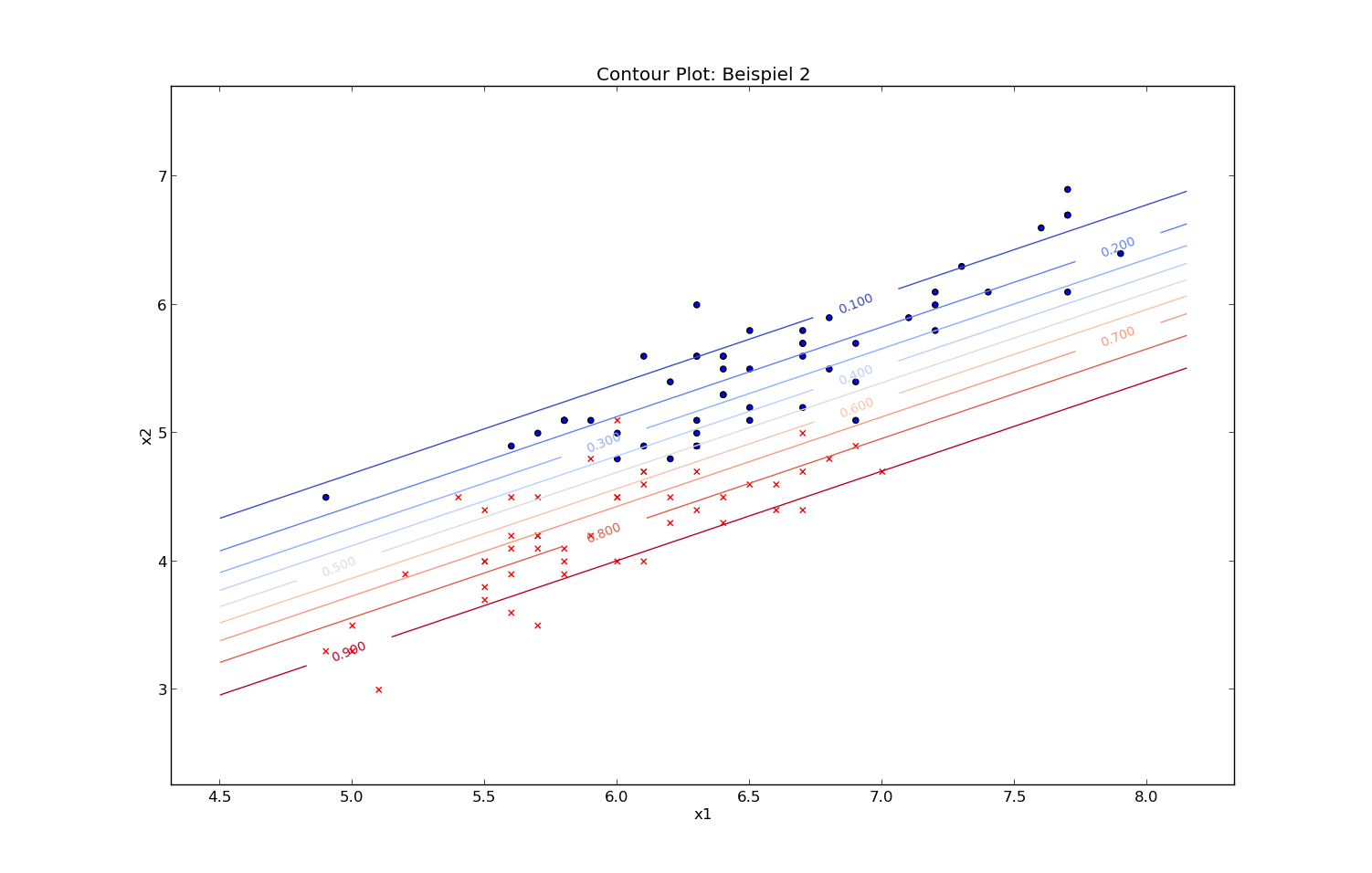
Probabilities (2d-plot) - Iris data:
Linear Separability¶
If we can separate the data points in n-dimensional space of two classes with a n-1 dimensional hyper plane than the classification problem is linearly separable.
Note: A hyper plane is formally defined by $\vec w^T \cdot \vec x = C$. So by definition logistic regression can solve only linearly separable data points completely ($C=-\theta_0$ and $w=(\theta_1, \dots, \theta_n)$).
# Generate training data
# Polar coordinates: r, phi
def get_x(nb, c):
assert (c==0 or c==1)
r = c + np.random.rand(nb)
phi = np.random.rand(nb) * 2 * np.pi
return np.concatenate(((r * np.sin(phi)).reshape(-1,1), (r * np.cos(phi)).reshape(-1,1)), axis=1)
def get_data(nb_1, nb_2):
x_0 = get_x(nb_1, 0)
x_1 = get_x(nb_2, 1)
X = np.concatenate((x_0, x_1), axis=0)
t = np.zeros(len(X))
t[len(x_0):] = 1
p = np.random.permutation(range(len(X)))
# permute the data
X = X[p]
t = t[p]
return X, t
X_train, t_train = get_data(30, 35)
X_test, t_test = get_data(20, 20)
def plot_train_data(x_0, x_1, circle=False):
plt.figure(figsize=(5,5))
plt.scatter(x_0[:,0], x_0[:,1], label='Class 0', color='b')
plt.scatter(x_1[:,0], x_1[:,1], label='Class 1', color='r')
plt.xlabel("$x_1$")
plt.ylabel("$x_2$")
if circle:
ra = np.arange(-1,1.01,0.01, dtype=np.float64)
ci = np.ndarray(len(ra))
ci[:-1] = (np.sqrt(1. - ra[:-1]**2))
ci[-1] = 0. # hack
plt.plot(ra, ci,'g-')
plt.plot(ra, -ci,'g-')
plt.xlim(-2.,2.)
plt.ylim(-2.,2.)
Example: non linear separable data¶
plot_train_data(X_train[t_train==0], X_train[t_train==1])
Any ideas, how we can extend the model?
plot_train_data(X_train[t_train==0], X_train[t_train==1], circle=True)
Non-linear basis functions $\phi({\bf x})$¶
Equation of a circle: $x_1^2 + x_2^2 = 1$}
Basis functions:
- $\phi_1({\bf x}) = x_1^2$
- $\phi_2({\bf x}) = x_2^2$
def plot_train_transformed(x_0, x_1):
plt.figure(figsize=(5,5))
plt.scatter(x_0[:,0], x_0[:,1], label='Class 0', color='b')
plt.scatter(x_1[:,0], x_1[:,1], label='Class 1', color='r')
plt.xlabel("$\phi_1($")
plt.ylabel("$\phi_2$")
ra = np.arange(-0.,1.,0.01)
plt.plot(ra, 1. - ra,'g-')
plt.xlim(-0.1,4)
plt.ylim(-0.1,4)
In the new feature space $\phi$ the data is linear separable:
phi_train = X_train**2
plot_train_transformed(phi_train[t_train==0], phi_train[t_train==1])
With proper basis function we can solve complex problem.
Cost function for classification¶
Recap: Problem formulation¶
- Train data $\mathcal{D}$: $\{ ({\vec x}^{(1)},y^{(1)}), ({\vec x}^{(2)},y^{(2)}), \dots ({\vec x}^{(m)},y^{(m)\
}) \}$
- ${\vec x}^{(i)} \in \mathbb{R}^{n+1}$, aa row vector $({\vec x}^{(i)})^T = (x_0,x_1, \dots, x_n)$ mit $x_0 =\ 1$
- $y \in \{0,1\} $
- Hypothesis: $h_{\theta}(x) = 1 / (1 + \exp\{- \vec \theta^T {\vec x}\})$
- We are looking for "good" parameters $\theta$
Loss¶
Recap: cost for linear regression $$ J(\theta) = \frac{1}{2m} \sum_{i=1}^m (h_\theta({\vec x}^{(i)})-y^{(i)})^2 $$
Loss (pointwise cost for a training example):
$$ J(\theta) = \frac{1}{m} \sum_{i=1}^m loss(h_\theta({\vec x}^{(i)}),y^{(i)}) $$
squared loss function (for linear regression):
$$ \text{loss}(h_\theta({\vec x}),y) = \frac{1}{2} (h_\theta({\vec x})-y)^2 $$
Loss for logistic regression¶
- quadratic loss (squared loss) is not used for logistic regression
- no convex problem
- from theorie ("maximum likelihood principle" ) we get another loss function.
Loss for an example of the positive class (y=1)¶
$\text{loss}(h_\theta(x^{(i)}), y^{(i)}) = -\log(h_\theta(x^{(i)}))$
z = np.arange(0.01,1.05,0.01)
plt.plot(z, -np.log(z)); plt.xlabel("z"); plt.ylabel("-log(z)"); plt.ylim(0,5); plt.xlim(0,1)
Loss for an example of the negative class (y=o)¶
$\text{loss}(h_\theta(x^{(i)}), y^{(i)}) = 1-\log(1- h_\theta(x^{(i)}))$
z = np.arange(0.0,1.0,0.01)
plt.plot(z, -np.log(1-z)); plt.xlabel("z"), plt.ylabel("-log(z)"); plt.ylim(0,5); plt.xlim(0,1)
Cross entropy¶
$$ \text{loss}(h_\theta(x^{(i)}), y^{(i)}) = - y^{(i)} \log (h_\theta(x^{(i)})) - (1 - y^{(i)}) \log \left( 1- h_\theta(x^{(i)})\right) $$\begin{align*} J(\theta) & = \frac{1}{m} \sum_{i=1}^{m} \text{loss}(h_\theta(x^{(i)}), y^{(i)}) \\ & = - \frac{1}{m} \left[ \sum_{i=1}^{m} y^{(i)} \log h_\theta(x^{(i)}) + (1 - y^{(i)}) \log \left( 1- h_\theta(x^{(i)})\right) \right] \end{align*}Gradient descent¶
Problem formulation¶
- Hypothesis: $h_{\theta}(x) = 1 / (1 + \exp\{- \theta^T {\vec x}\})$
- Cost function:
Goal: "good" parameters $\theta$ for the hypothesis $h_\theta({\bf x})$ by $\text{minimize}_{\theta} J(\theta)$
Update Rule for all $\theta_j$ (gradient descent) $$ \theta_j^{old} \leftarrow \theta_j^{old} - \alpha \frac{\partial}{\partial \theta_j} J(\theta^{old}) $$
So we get for the Update Rule:
$$ \theta_j^{new} \leftarrow \theta_j^{old} - \alpha \frac{1}{m} \sum_{i=1}^{m} \left( h_\theta({\vec x}^{(i)})- y^{(i)} \right) x_j^{(i)} $$Exercise: Implementation of logistic regression¶
Note: Modify your "multivariate linear regression code", that your implementation works for linear and logistic regression.
Implement the logistic regression model as python function:
get_logistic_hypothesis(theta)The python function should return a function, e.g.
>> theta = np.array([1.1, 2.0, -.9]) >> h = get_logistic_hypothesis(theta) >> print h(X) array([ -0.89896965, 0.71147926, ....Implement (as function)
- the cross-entropy loss for logistic regression (in general for classification) and
- the squared-loss for linear regression.
The function should return a function, e.g.
>> loss = get_cross_entropy_loss(h, X, y) >> print loss(theta) array([ 7.3, 9.5, ....The array has m elements (number of data examples, i.e. a loss value for each example)
Note: if we pass as argumenththe function could be used for any classification problem (resp. regression problem.)Implement the cost function:
>> cost_function = get_cost_function(loss)lossandcost_functionare functions with argumenttheta.>> cost_function(theta) 15525.3Implement gradient descent by using the functions above.
Implement (modify) a function which does the update rule for computation of the new $\theta$ values:
theta = compute_new_theta(x, y, theta, alpha, hypothesis)
- Implement (or modify) a function
gradient_descent(alpha, theta, X, y).thetaare the start values for $\vec \theta$. The function applies iterativelycompute_new_theta. Use an arbitrary stoping criterion.
Use the artifical data (see below) for learning a logistic regression model. Plot the progress (cost value over iterations).
Plot the decision boundary together with the data scatter plot.
Hint: $\theta_0 + \theta_1 x_1 + \theta_2 x_2 = 0$Implement the computation of the classification error (one-line in numpy, no python loop!).
classification error = number of wrong classified examples / number examples
# Artifical data
# class 0:
# covariance matrix and mean
cov0 = np.array([[5,-3],[-3,3]])
mean0 = np.array([2.,3])
# number of data points
m0 = 1000
# generate m0 gaussian distributed data points with
# mean0 and cov0.
r0 = np.random.multivariate_normal(mean0, cov0, m0)
# covariance matrix
cov1 = np.array([[5,-3],[-3,3]])
mean1 = np.array([1.,1])
m1 = 1000
r1 = np.random.multivariate_normal(mean1, cov1, m1)
# class 0:
# covariance matrix and mean
cov0 = np.array([[5,-4],[-4,4]])
mean0 = np.array([2.,3])
# number of data points
m0 = 1000
# class 1
# covariance matrix
cov1 = np.array([[5,-3],[-3,3]])
mean1 = np.array([1.,1])
m1 = 1000
# generate m0 gaussian distributed data points with
# mean0 and cov0.
r0 = np.random.multivariate_normal(mean0, cov0, m0)
r1 = np.random.multivariate_normal(mean1, cov1, m1)
plt.scatter(r0[...,0], r0[...,1], c='b', marker='o', label="class 0")
plt.scatter(r1[...,0], r1[...,1], c='r', marker='x', label="class 1")
plt.xlabel("$x_1$")
plt.ylabel("$x_2$")
plt.legend()
plt.show()
X = np.concatenate((r0,r1))
y = np.zeros(len(r0)+len(r1))
y[:len(r0),] = 1
Multiclass problems¶
Multiclass problems¶
by using binary classifier:
One vs. All (One vs. Rest):
- Train $k$ binary classifier $h^{(u)}$ for each class $u$.
- Classification by $h = max_u h^{(u)}(x)$
All vs. All (One vs. One or All-Pairs):
- Train $k(k-1)$ binary classifier $h^{(uv)}$ for each pair of the classes $u$ and $v$ (Note $h^{(uv)}=h^{(vu)}$).
- Classification by $h = max_u \sum_v h^{(uv)}(x)$
Softmax¶
One-hot output encoding
- for each class $i$ one Output $y_j$ with the target value
- $y_j = 1$, if the train example belongs to class $j$
- $y_j = 0$, else
- for each class $k$ a parameter vector $\vec{\theta}_{k}$ (e.g. as matrix)
- for each class $i$ one Output $y_j$ with the target value
Prediction of the probability that a data set with input vector $\vec{x}$ belongs to class k: $$ P(y=k \mid \vec x, \vec \theta_{k}) = softmax(\vec \theta_k \cdot \vec x) = \frac{e^{\vec \theta_k \cdot \vec x}}{\sum_j e^{\vec \theta_j \cdot \vec x}} $$
Class prediction by $$ y_{pred} = argmax_k P(y=k \mid {\vec x}, \theta^{(k)}) $$
Literature¶
- Andrew Ng: Machine Learning. Openclassroom Stanford University, 2013
- C. Bishop: Pattern recognition and Machine Learning, Springer Verlag 2006
- Video: Nando de Freitas: Machine Learning, Logistic Regression
- M. Mohri et.al.: Foundation of machine learning, 2012